Featured Items

The most promising strategies to defeat antibiotic-resistant bacteria

Antibiotic resistance intitiative at isglobal*
updated last: 11 november 2016
Much has been said and written these last years on the alarming rise worldwide in the number of bacteria of medical importance that are resistant to one or more antibiotics as a result of antibiotic abuse and misuse, not only in humans but also in agriculture: in the last 30 years, antibiotic use in farm animals has increased between 600% (penicillin-related drugs) and 1,500% (tetracyclines).
To a certain extent, the problem of antimicrobial resistance is similar to that of climate change: it is the result of irresponsible human behaviour and can have drastic consequences for mankind. However, as with climate change, we have the knowledge and tools to tackle the problem and win the war against antimicrobial resistance.
In this digital report we’d like to highlight what is being done and achieved in four key battlefronts against resistant bacteria: the search for new antibiotics, advances in microbiological diagnosis, the study of resistance mechanisms, and antimicrobial surveillance.
1. New Weapons
(New Antibiotics)
Why is it a key battleground?
To combat antimicrobial resistance, we need new antibiotics and we need them fast. And despite this, the antibiotic development pipeline of the major pharmaceutical companies has basically dried up. Not one single new antibiotic class has been discovered since 1987, and out of the 8 antibiotics approved by the Food and Drug Administration (FDA) agency, most do not have new mechanisms of action or are simply combinations of existing antibiotics. It is as if in the 21st century we used bayonets to combat an army equipped with guided missiles.
What tactics are being used?
a) Adapt the available weapons
One approach to treat infections by drug-resistant bacteria is the revival of forgotten antibiotics such as colistin, in order to use them correctly and to their full potential and “alleviate” the use of more recent antibiotics, as well as alternating antibiotics that are frequently used in hospital settings. Nevertheless, this is not a solution in the long-term since resistant strains will inevitably emerge.
Another approach that is being tested is the combination of different antibiotics that act through different pathways. For example, the combination of beta lactams and clavulanic acid (an inhibitor of the bacterial enzyme beta lactamase) has been successful. However, beta lactamase variants resistant to inhibition by clavulanic acid have started to emerge. Available antibiotics can also be modified in order to increase their activity and overcome resistance mechanisms, but it is difficult to alter the structure of a drug without rendering it inactive.
b) Identify new drug targets
New weapons can be developed if new targets (i.e. bacterial molecules or metabolic pathways that can be inhibited) are identified. Bacteria express around 200 key conserved proteins but the number of targets exploited by existing drugs is extremely low. Thanks to bacterial genome-sequencing projects and to bioinformatic tools, it is possible to identify new antibiotic targets in silico (i.e. by computer simulation). For example, the comparison of the genome of Streptococcus pneumoniae with a related free-life bacterium (S. thermophilus) has allowed the identification of two pathogenic genes (expressed by the former but not by the latter) that can be potentially inhibited by drugs.
c) Search for new weapons in nature
To date, no antibody on the market comes from the sea despite the fact that it represents the most important – and unexplored- source of natural products in our planet. Algae, sponges and microorganisms living at the bottom of the sea are only some examples of organisms from which compounds with antimicrobial activity have already been isolated. For example, a bacterium (Streptomyces sp) collected near the Santa Barbara coast in California produces anthracimycin, a molecule that can kill methicillin-resistant Stapylococcus aureus (MRSA).
NoMorFilm, searching for antibiotics in the sea
At ISGlobal we are leading the European project NoMorFilm, that will screen marine microalgae in search for new molecules with activity against bacteria and biofilms (bacterial films that are very difficult to treat with available antibodies).
Free-living bacteria, particularly those living in the soil or in hostile environments, represent another rich but poorly explored source of antimicrobial compounds. Many of them are difficult to cultivate in the laboratory, but recently developed techniques such as DNA transfer and metagenomics are allowing the sequencing of their entire genome and the identification of candidate molecules with antibacterial activity.
Our body also harbors a large quantity of bacteria, virus and fungi. In our gut alone we have trillions of bacteria (more than 1,000 different species and 3 million genes). This microbiota is not only harmless, but it can also defend us against pathogenic bacteria. Thus, a Staphylococcus that normally inhabits our nose secretes a peptide (called lugdunin) that inhibits colonization by the pathogenic Staphylococcus aureus.
Nature offers numerous options to fight antibiotic-resistant bacteria, such as natural products derived from the sea, bacteria living in hostile environments, our own microbiota, and bacteriophages.
The need for new antimicrobial agents has revived an interest in bacteriophages, up to now mainly studied and used in ex-USSR and Eastern Europe. These viruses specifically infect and destroy bacteria, and have been shown to efficiently destroy biofilms, at least in the laboratory. Bacteriophages can be used in conjunction with other antibiotics: for instance, the phage OMKO1 binds to the cell membrane where the bacterium Pseudomonas aeruginosa pumps out antibiotics, making it more vulnerable these drugs. The problem is that phage therapy requires knowing what bacterium is causing the infection. In addition, the production of stable phage lots is complicated and their efficacy in treating human infections remains to be demonstrated with controlled clinical trials.
Promising natural peptides (small aminoacidic chains) have been isolated from different organisms in nature. For instance, endolysins, derived from bacteriophages, degrade the bacterial cell wall and have proved to be efficient in treating skin infections in clinical trials. At ISGlobal, we have isolated six peptides from frog skin that have activity against colistin-resistant bacteria.
Highly promising results have been obtained with fecal microbiota transfers from healthy donors to patients with recurrent infections by 'Clostridium difficile'
An alternative to killing pathogenic bacteria is “disarming” them in order to make them less aggressive. To do so, it is necessary to find molecules that can interfere for example with the secretion of bacterial toxins, or with host infectivity. For instance, the so-called pilicides interfere with the formation of pilli, a structure used by E. coli to attach to the host cell. Since antivirulence molecules do not kill the bacterium, they do not exert such a strong selective pressure as compared to antibiotics and therefore have the great advantage of slowing down the development of resistance mechanisms. Interestingly, if combined with antibiotics, they could also help decrease the effective doses. Although a promising strategy, antivirulence molecules have not yet reached the clinical trial phase.
Finally, a completely different strategy consists in replacing whole populations of bacteria. Fecal microbiota transplants (from a healthy donor) have yielded highly promising results in the treatment of patients with recurrent infections by Clostridium difficile, a bacterium that causes severe diarrhea.
2. Reconnaissance
(diagnosis)
Why is it crucial and what is needed?
Just like in a battle it is important to obtain information of the enemy (troops, resources, weapons), in the war against antimicrobial resistance it is essential to know, as quickly as possible, what type of bacteria and resistance mechanisms we are dealing with in order to choose the adequate treatment. Normally, the traditional bacterial culture techniques require between 48 and 72 hours to isolate and identify the pathogenic organism. In contrast, real-time detection permits adapting immediately the treatment to the bacteria concerned. This not only results in a better treatment of the patient but also reduces the unnecessary use of wide-spectrum antibiotics that promote resistance.
A diagnosis in real time permits choosing the right treatment for the patient from the beginning and reducing the unnecessary use of antibiotics
In addition to being rapid, the ideal diagnostic technique needs to be sensitive (detect the disease-causing bacterium even when at low numbers) and specific (distinguish it from other similar bacteria); inform on the presence and type of resistance genes; be feasible in low and middle-income countries (without requiring highly specialized personnel or equipment); and be affordable.
What is being done?
The detection of bacterial DNA by means of its amplification by PCR (polymerase chain reaction) is one of the diagnostic tools which has most impacted microbiological diagnosis, particularly in terms of speed – in a routine laboratory, the time needed to identify the pathogen and detect possible antibiotic-resistant genes can be reduced to less than two hours.
As an example, in ISGlobal we have set up a rapid detection assay of the main antibiotic-resistant genes in gram-negative bacteria- extended spectrum beta-lactamases and carbapenases- directly from blood cultures, and in only 15 minutes. We have also managed to detect, taking blood from a catheter, the presence of MRSA in 90 minutes.
The main challenge for DNA amplification assays is the relatively limited range of resistance mechanisms that can be detected. Another problem is that, even when a resistance gene is detected, its level of expression may not be high enough to translate into a functional resistance.
An emerging and highly promising technique is the so-called MALDI-TOF/MS that allows the quick identification of different bacterial species and can even distinguish between different strains of the same species. The method is based mass spectrometry, which identifies a bacterium’s set of proteins according to its mass. In ISGlobal we have developed an automated MALDI-TOF protocol for the categorization of clinical isolates of meticilin-resistant Staphylococcus aureus in a matter of minutes. Importantly, the technique can be used in routine laboratories that already use mass spectrometry for bacterial identification.
Nonetheless, there is still some way to go before these techniques are implemented in low and middle-income countries: commercial platforms and kits that reduce the need for highly trained personnel but that are accessible and affordable, need to be developed.
3. The enemy’s strategies
(mechanisms of antibiotic resistance)
Why is it a key battlefield?
Understanding the mechanisms by which bacteria defend themselves against an antibiotic aggression is key to finding ways of increasing the efficacy and half-life of available antibiotics, as well as delaying the development of resistance to new drugs.
A bacterium can defend itself from an antibiotic “attack” through four general mechanisms:
a) Reducing the number of channels that antibiotics use to enter the cell. A decrease in the number of channels (named porins) is associated with resistance to quinolones.
b) Degrading or inactivating the antibacterial drug once it is inside, thanks to the action of proteins called enzymes. Bacterial beta-lactamases represent one of the main mechanisms of resistance to beta-lactams, widely used antibiotics that inhibit cell wall synthesis.
c) Modifying the molecule or metabolic pathway targeted by the drug: vancomycin-resistant bacteria have an alteration in the site recognized by the antibiotic, rendering it useless.
d) Pumping the antibiotic out of the cell, thanks to bacterial efflux pumps located in the cell membrane. This is one of the main mechanisms used by Pseudomonas aeruginosa to defend itself from the action of aminoglycosides.
In turn, bacteria can acquire antibiotic-resistant genes through the following mechanisms:
a) Spontaneous mutations in their DNA
b) Transfer of resistant genes from one bacteria to the other via:
- Transformation: the bacterium captures DNA from the environment
- Conjugation: plasmids (small DNA circles) that contain resistance genes may be passed from one bacterium to the other. This horizontal transfer of genes is worrying since one single plasmid can harbor several resistance genes. In 1968, for instance, 12,500 persons in Guatemala died of a diarrhea epidemic caused by Shigella. The bacterium contained a plasmid with genes that conferred resistance to four antibiotics. In USA, a woman suffered a urinary infection by an E. coli strain that contained 15 antibiotic-resistant genes on two plasmids. One of those genes was mcr-1, which confers resistance to colistin, a last resort antibiotic.
- Transduction: resistance genes are transferred from one bacterium to another via bacteriophages (virus that infect bacteria).
What is being done?
On one hand, the mechanisms and consequences of antibiotic resistance need to be further characterized, particularly for those bacteria that are a leading cause of hospital acquired infections. Interestingly, at ISGlobal we have found that when the hospital-associated pathogen Acinetobacter nosocomialis acquires resistance to colistin, it suffers changes in its external membrane composition and decreases its virulence. Similarly, quinolone resistance in Salmonella enterica reduces the expression of virulence-associated genes. These results, although reassuring, are not generalizable.
It is important to understand how antibiotic resistance develops, as well as to foresee future resistance mechanisms
Understanding the path that leads to the development of antibiotic resistance is another key aspect. For instance, we have analysed the evolution of resistance mechanisms in Pseudomonas aeruginosa isolated from patients in the intensive care unit, and show that the stepwise acquisition of several mechanisms is needed to reach a “breakpoint” beyond which susceptibility to drugs is lost. When Streptococcus pneumoniae is submitted to increasing antibiotic doses, the immediate response requires mutations in a small number of genes (less than 10) but the problem is that two bacteria can reach the same results (survival and growth) using different genes.
That is why it is also important to go one step further and foresee future resistance mechanisms. There is growing evidence that antibiotic-resistant genes are widely distributed and that many ecosystems related to agriculture such as soil, residual waters or animal manure are natural reservoirs of potential resistance genes. Metagenomic studies performed on these reservoirs start to provide valuable insight on new resistance genes that may emerge. For instance, a study performed with meticilin resistant Staphylococcus aureus isolated from recreational beaches or high contact surfaces in built environments showed that 98% of the bacteria analysed were resistant to other antibiotics, reflecting the antibiotic-resistant “gene pool” in the environment.
4. The Secret Service
(surveillance)
Why is this critical?
It is often said that, on the long run, a war is won behind the battle lines thanks to information and intelligence services. The same applies to the war against antimicrobial resistance. Surveillance studies provide valuable information to detect changing trends in the incidence of infectious microorganisms in humans and of resistance mechanisms, at a national and global level, as well as to guide specific and adequate interventions according to each context. In addition, comparison of data over time allows evaluating the impact of such interventions.
What is being done?
Surveillance implies monitoring and detecting the appearance of new resistant strains. In this sense, at ISGlobal we have managed to trace the spread of a carbapenem-resistance gene (OXA-23) in the bacterium A. baumannii across the Mediterranean region. Moreover, we have characterized one of the first A. pitti strains bearing the NDM-1 gene (a beta lactamase first described in New Delhi and that confers resistance to carbapenems), isolated from a patient in Turkey. The results suggest that the NDM gene circulated among other Acinetobacter species before spreading to species of medical relevance.
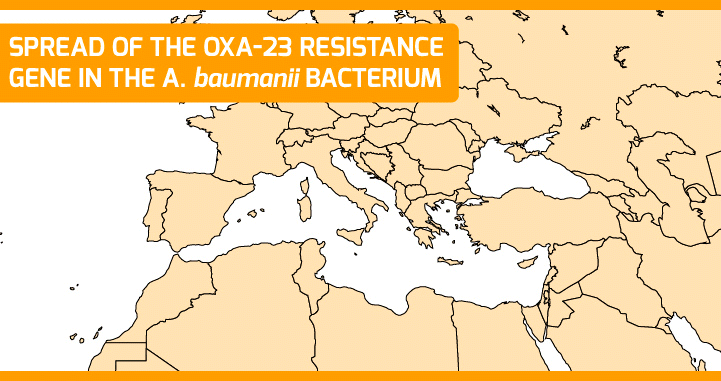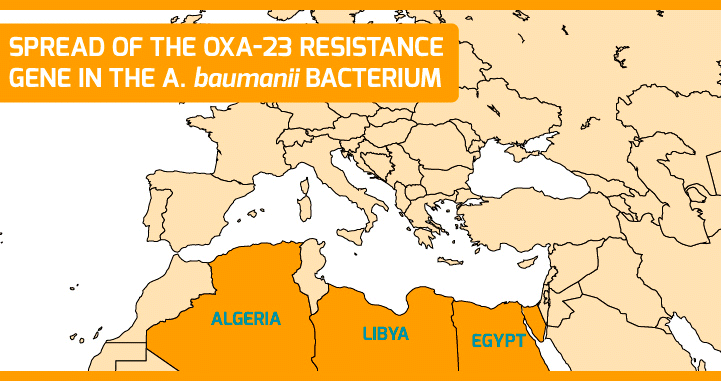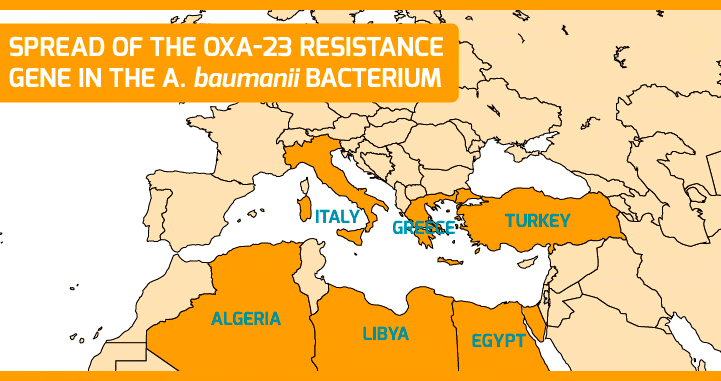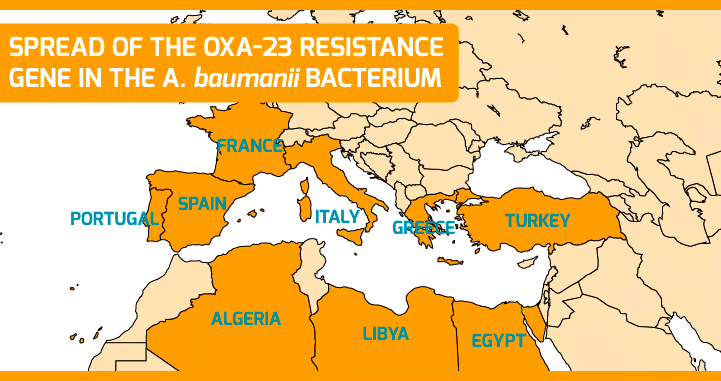
Surveillance of antimicrobial resistance lacks coordination and harmonization at the global level
Studies like the above, performed in numerous hospitals and health centres, feed data into national and regional projects and networks that collect information on the prevalence of antibiotic resistance genes. In Europe, EARS-Net is a network of national surveillance systems that collects data from 900 laboratories that provide service to more than 1,400 hospitals (and 100 million European citizens). The network surveys antimicrobial susceptibility of seven bacterial pathogens that cause the most infections in humans (Streptococcus pneumonia, Staphylococcus aureus, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella pneumonia, Pseudomonas auruginosa).
Many of these networks also provide data on the types and doses of prescribed antibiotics, which permits correlating the use of certain antibiotics with the increase or decrease in certain resistance genes and in turn design protocols that slow down their emergence.
Nevertheless, as the WHO warns in its 2014 report, surveillance of antimicrobial resistance lacks coordination and harmonization at the global level, and there are significant gaps in information on pathogens of major public health importance (such as multidrug-resistant tuberculosis).
Conclusion
The emergence of genes that can confer resistance to different antibiotics is something we cannot avoid – it’s part of the natural evolution process of any microorganism. However, we can avoid selecting the bacteria that express such genes as a result of the abuse and misuse of current and future antibiotics.
The good news is that we have the knowledge, tools and promising strategies to defeat, or at least control, the emergence and spread of multiresistant bacteria in the near future. Bacteria have a large number of potential drug targets that we have not yet exploited. The sea and other natural resources are an incredibly rich and diverse source of products with antibiotic potential as a result of millions of years of natural selection. The development of new diagnosis techniques allows us to adapt ever more rapidly the adequate treatment for each patient and avoid the unnecessary use of antibiotics. We are starting to understand the mechanisms that lead to the emergence and selection of resistance mechanisms and their consequences. And last, but not least, we are building and strengthening networks to survey the spread of antimicrobial resistance.
Scientific research is, therefore, an essential component in the war against antibiotic resistance. But it is not sufficient. Raising awareness among the population and implementing laws that guarantee the rational use of antibiotics (also in the food sector) and provide incentives for the development of new drugs is also required. Without this, no matter how many soldiers and weapons we have, we will not win the battle.












